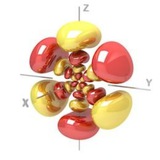
Exotic bond takes computational chemists by surprise | Research | Chemistry World
https://www.chemistryworld.com/news/exotic-bond-takes-computational-chemists-by-surprise/4013547.article
https://www.chemistryworld.com/news/exotic-bond-takes-computational-chemists-by-surprise/4013547.article
Chemistry World
Exotic bond takes computational chemists by surprise
New results could help to solve the dispute around the strange Na-B bond in NaBH3-
Atomic_and_electronic_structure_of_solids_by_Efthimios_Kaxiras_z.pdf
7.1 MB
Atomic and Electronic Structure of Solids - Efthimios Kaxiras
Chemical code used to store Jane Austen quote in plastic molecules | New Scientist
https://www.newscientist.com/article/2275136-chemical-code-used-to-store-jane-austen-quote-in-plastic-molecules/
https://www.newscientist.com/article/2275136-chemical-code-used-to-store-jane-austen-quote-in-plastic-molecules/
New Scientist
Chemical code used to store Jane Austen quote in plastic molecules
It is now possible to store information in the atomic structure of plastic molecules using a hexadecimal code that allows for greater storage density than a binary system
Mathematical physics in theoretical chemistry by Blinder.pdf
3.2 MB
Mathematical Physics in Theoretical Chemistry
A new release of MayaChemTools, an open source collection of command line
noscripts, is now available containing the following Python noscripts based on
Psi4:
o Psi4CalculateEnergy.py
o Psi4CalculatePartialCharges.py
o Psi4CalculateProperties.py
o Psi4GenerateConformers.py
o Psi4PerformMinimization.py
o Psi4VisualizeDualDenoscriptors.py
o Psi4VisualizeElectrostaticPotential.py
o Psi4VisualizeFrontierOrbitals.py
These Python noscripts rely on the availability of Psi4 and RDKit in your
environment. The RDKit is used to process molecules and generate initial 3D
coordinates for Psi4.
http://www.mayachemtools.org/
noscripts, is now available containing the following Python noscripts based on
Psi4:
o Psi4CalculateEnergy.py
o Psi4CalculatePartialCharges.py
o Psi4CalculateProperties.py
o Psi4GenerateConformers.py
o Psi4PerformMinimization.py
o Psi4VisualizeDualDenoscriptors.py
o Psi4VisualizeElectrostaticPotential.py
o Psi4VisualizeFrontierOrbitals.py
These Python noscripts rely on the availability of Psi4 and RDKit in your
environment. The RDKit is used to process molecules and generate initial 3D
coordinates for Psi4.
http://www.mayachemtools.org/
👍1
Physicists Harnessed Thousands of Molecules Into a Single Quantum State
https://www.sciencealert.com/scientists-have-harnessed-thousands-of-molecules-into-a-single-quantum-state
https://www.sciencealert.com/scientists-have-harnessed-thousands-of-molecules-into-a-single-quantum-state
ScienceAlert
Physicists Harnessed Thousands of Molecules Into a Single Quantum State
In a major milestone for quantum physics, thousands of molecules have been induced to share the same quantum state, dancing together in unison like one huge super molecule.
Physical_Inorganic_Chemistry_A_Coordination_Chemistry_Approach_KETTLE.pdf
40.1 MB
Physical Inorganic Chemistry: A Coordination Chemistry Approach - KETTLE
Essentials_of_coordination_chemistry_a_simplified_approach_with.pdf
17.8 MB
Essentials of coordination chemistry : a simplified approach with 3D visuals - Bhatt, Vasishta
Ligand Overlay enables drug discovery without the target protein structure. Learn to use it in your ligand-based research workflow and how the CCDC uses agile development Thurs., 20 May. Click to register.Down pointing backhand index
#pharma #DrugDiscovery #chemistry
https://register.gotowebinar.com/register/6914250695137108238
#pharma #DrugDiscovery #chemistry
https://register.gotowebinar.com/register/6914250695137108238
EMSL arrows is a scientific service that uses NWChem and chemical computational databases to make materials and chemical modeling accessible via a broad spectrum of digital communications including posts to web APIs, social networks, and traditional email. Molecular modeling software has previously been extremely complex, making it prohibitive to all but experts in the field, yet even experts can struggle to perform calculations. This service is designed to be used by experts and non-experts alike. Experts will be able carry out and keep track of large numbers of complex calculations with diverse levels of theories present in their workflows. Additionally, due to a streamlined and easy-to-use input, non-experts will be able to carry out a wide variety of molecular modeling calculations previously not accessible to them.
You do not need to be a molecular modeling expert to use EMSL Arrows. It is very easy to use. You simply email chemical reactions to arrows@emsl.pnnl.gov and then an email is sent back to you with thermodynamic, reaction pathway (kinetic), spectroscopy, and other results. There are currently 42,000+ calculations in the EMSL Arrows database and it is growing every day. If an EMSL Arrows request requires a calculation not already in the database, then it will automatically start the calculation on a small number of freely available computers and send back the results when finished. More information can be found at Arrows. We would like thank the DOD SERDP program and the DOE OS OBER EMSL project for their support.
https://nwchemgit.github.io/Current_events.html#emsl-arrows-an-easier-way-to-use-nwchem
You do not need to be a molecular modeling expert to use EMSL Arrows. It is very easy to use. You simply email chemical reactions to arrows@emsl.pnnl.gov and then an email is sent back to you with thermodynamic, reaction pathway (kinetic), spectroscopy, and other results. There are currently 42,000+ calculations in the EMSL Arrows database and it is growing every day. If an EMSL Arrows request requires a calculation not already in the database, then it will automatically start the calculation on a small number of freely available computers and send back the results when finished. More information can be found at Arrows. We would like thank the DOD SERDP program and the DOE OS OBER EMSL project for their support.
https://nwchemgit.github.io/Current_events.html#emsl-arrows-an-easier-way-to-use-nwchem
Fundamentals_of_molecular_symmetry_by_Jensen,_Per_Bunker,_Philip.pdf
23.8 MB
Fundamentals of molecular symmetry by Jensen, Per Bunker, Philip R.
Building molecules like Tinkertoys? A breakthrough study may pave the way | University of Chicago News
https://news.uchicago.edu/story/building-molecules-tinkertoys-breakthrough-study-may-pave-way
https://news.uchicago.edu/story/building-molecules-tinkertoys-breakthrough-study-may-pave-way
University of Chicago News
Building molecules like Tinkertoys? A breakthrough study may pave the way
UChicago chemists aim to transform the field of chemical discovery
Electrons_in_Molecules_From_Basic_Principles_to_Molecular_Electronics.pdf
19.7 MB
Electrons in Molecules From Basic Principles to Molecular Electronics by Jean-Pierre Launay, Michel Verdaguer
Artificial Intelligence with Python.pdf
34.7 MB
Artificial Intelligence with Python